1. Nanopore Project
2. Functional Genomics
3. Collaborations
Experimental and computational functional genomics in model organisms such as yeast  Saccharomyces cerevisiae , fruit fly Saccharomyces cerevisiae , fruit fly  Drosophila melanogaster , nematode Drosophila melanogaster , nematode  Caenorhabditis elegans , plant Caenorhabditis elegans , plant  Arabidopsis thaliana , mouse Arabidopsis thaliana , mouse  Mus musculus , and in human Mus musculus , and in human  cells. cells.
- Identification of DNA sequence and RNA expression variation in any organism, using custom
synthesized high-density oligonucleotide arrays
 . .
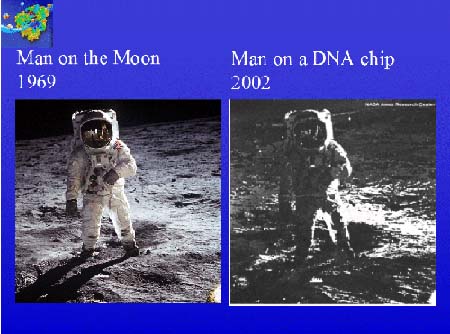
This image demonstrates the inherent flexibility of high-density oligonucleotide arrays produced by the maskless procedure. To produce this image, a photograph of an astronaut on the surface of the Moon from 1969 was digitized and converted to a four-level gray-scale image. A distinct 18mer oligonucleotide probe corresponding to perfect match sequence - or one, two, or three mismatches - was assigned to each of the four gray-scale values. Virtual masks were then designed using this information and an array was synthesized using the Maskless Array Synthesizer (MAS) system. After synthesis, the array was hybridized with a single biotinylated oligonucleotide, which has the perfect match sequence, stained with streptavidin-cy3. This demonstration illustrates the versatility of the high-density DNA oligonucleotide array synthesis.
|
- Bio-informatics infrastructure for analysis of DNA sequence variation.
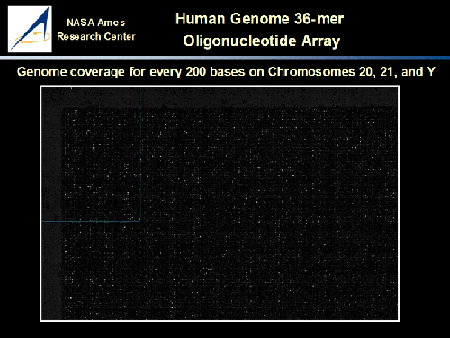
High-density oligonucleotide arrays have been custom designed, in situ synthesized and used in a pilot project for transcriptional profiling of the human genome. The design contained 180,000 features per array, density coverage for every 200 bases of the human chromosomes 20, 21, and 22 on two chips. The image represents a subset of hybridization signals on the array that was hybridized with total RNA isolated from human placenta.
|
- Functional genomics of the yeast gene-deletion strains
 to determine gene functions under any condition. to determine gene functions under any condition.
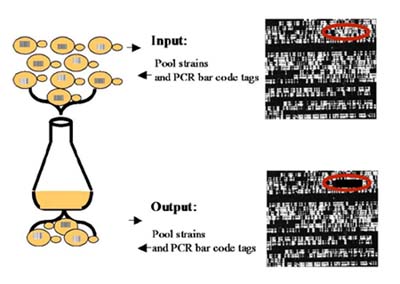
Disruption of gene function by mutation allows elucidation of both qualitative and quantitative functions of genes. Complete deletion of the entire coding sequence of a gene in yeast Saccharomyces cerevisiae results in measurable effects under specific growth conditions that affect the function of the deleted gene product. This information can be obtained without prior knowledge of specific effects of growth conditions on gene functions. Measurement of the relative growth rate of each yeast gene-deletion strain, over a period of several cell division generations, can be used to identify both qualitative and quantitative functions of each gene deletion mutation. This measurement is possible when the deleted genes are tagged in the yeast genome with unique 20-mer base 'bar-code' tags. Tag sequence complements are synthesized on the surface of the oligonucleotide array, to detect the quantitative phenotype of each yeast gene in a pool of all gene-deletion yeast mutants.
|
|
|

 Open the external link in a new window.
Open the external link in a new window.
 Show the picture in this window.
Show the picture in this window.